Drugs targeting RNA-protein interactions are part of the next generation of medicines, but the lack of predictive and cost-effective cell-based high-throughput screening assays has been a significant barrier to their development.
Synsight’s automated MTBench® platform is changing the game and opening exciting avenues in the field of RNA-targeting drug discovery.
Technological limits challenge RNA targeting drug discovery
RNA-Protein interactions are crucial for controlling gene expression, and their dysregulation can lead to many diseases, making them prime targets for drug development. Human cells express approximatively 3000 transcripts that encode RNA-Binding Proteins (RBPs), each potentially representing medically significant RNA-protein interactions.
Developing drugs that affect these interactions could represent a significant advancement for human health, but is severely underexplored. There are only a few published studies in this area (1) due to challenges in choosing the right chemicals from predictive in silico models and validating them experimentally. Choosing the right chemicals from computer models is hindered by the complexity of RNA structures and the intricate nature of RNA:protein interactions. Validating computationally predicted modulators at the laboratory workbench is even more difficult, as there is a need for innovative methods that can assess molecular interactions within the dynamic cellular environment. Even though some promising proof-of-concept RNA:protein interaction inhibitor assays have been recently reported (1), more work is needed to adapt these methods for high-throughput screening. Overcoming these challenges is vital for progress in RNA-targeting drug discovery.
MTBench® HTS assay identifies drug candidates
for RNA-targeting drug discovery
Synsight is developing an exclusive, patented technology to detect in cellulo interactions: the Microtubule Bench assay: MTBench®, originating from INSERM (French National Insitute of Health) (2). The MTBench® assay uses the microtubule network as an intracellular bench to selectively concentrate and detect RNA-protein interactions in living cells.
Since its initial introduction in 2015, Synsight has miniaturized and standardized the MTBench® technology through the application of lab automation and robotics (2), and has integrated it into its drug discovery process (3). This integration incorporates our advanced computer-aided drug design algorithms and screening cascades. Successful MTBench® miniaturization translates into optimal screening windows which are characterized by validated robustness scores, showing high repetitivity and reproducibility.
ZOOM ON MTBench®
the right assay to decipher RNA:protein interactions
To target interactions between mRNA and RNA-binding proteins, MTBench® requires the fusion of a fluorescently tagged RNA-binding protein of interest with a microtubule-binding domain (MBD). When expressed in a cell, the fusion protein is directed onto the microtubules, where it can behave like a bait and bind prey mRNA.
HOW DOES IT WORK ?
To target interactions between mRNA and RNA-binding proteins, MTBench® requires the fusion of a fluorescently tagged RNA-binding protein of interest with a microtubule-binding domain (MBD). When expressed in a cell, the fusion protein is directed onto the microtubules, where it can behave like a bait and bind prey mRNA. The MTBench® assay employs high-throughput microscopy to derive a correlation score from a single test condition, reflecting the colocalization between the fluorescence of baits and preys on the same microtubules. This score is then translated into a mathematical regression slope, serving as the assay output. In MTBench®, a positive slope indicates an interaction, while a reduced slope value signifies the inhibition of this specific interaction in the presence of an active compound.
This straightforward method allows for easy adaptation to target other interfaces,
like interactions between proteins or between a protein and a specific RNA sequence.
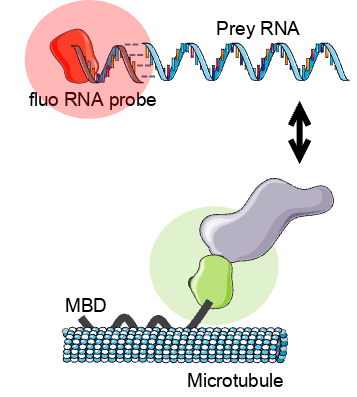
Methodological scheme of the MTBench® assay.
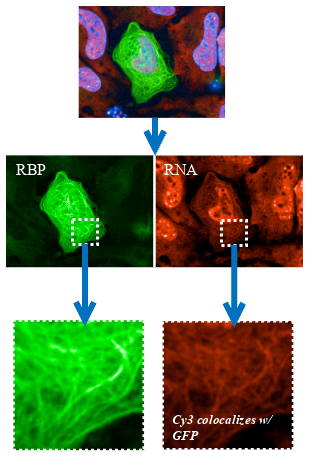
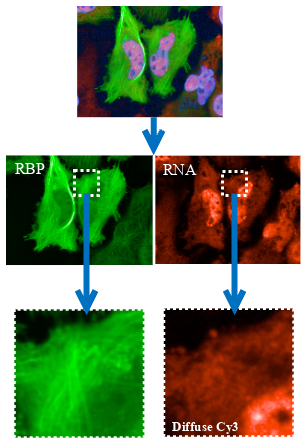
A fluorescent fusion RNA-binding protein, serving as a bait, is guided onto microtubules. In the absence of a pharmaceutical agent, the bait can interact with prey intracellular RNAs labeled with fluorescent oligos.

Representative images of HeLa cells are captured with our high-content imager, showcasing the colocalization of GFP (RNA-binding protein) and Cy3 (mRNA) signals in the absence or presence of a therapeutic agent.
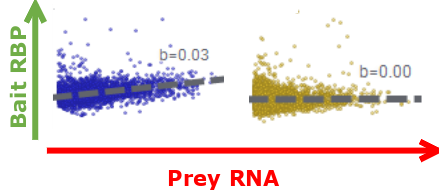
This graph below depicts the fluorescence intensities of prey and bait.
The slope value indicates the level of interaction between prey and bait,
with a null slope indicating an absence of interaction.
RELEVANCE AND SPECIFICITY COMPATIBLE WITH HIGH-THROUGHPUT
Assay relevance and specificity are ensured by systematic quality controls that include compound toxicity and selectivity. With a highly competitive cost per test data point and an experimental setup compatible with High-Throughput Screening, the MTBench® represents an innovative in cellulo screening assay opening the way to physiologically relevant RNA targeting in drug discovery.
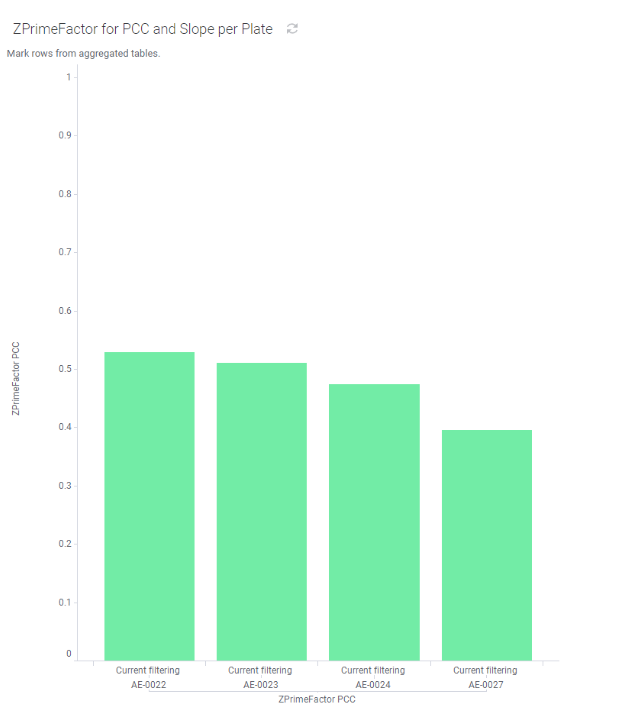
As described in the graphs below, our assay shows illustrating notable reproducibility, resulting in satisfactory Z’ robustness scores across four independent experiments conducted in two demonstration screening runs.
Application of MTBench® is compatible with Automated, High-Throughput Screening of RNA:YB1 Inhibitors.

The automated, microplate-based MTBench® platform developed by Synsight offers:
a high-throughput,
cost-efficient,
predictive,
and flexible solution for RNA-targeting drug discovery.
Integrated into the iterative Synsight drug discovery process,
MTBench® represents a notable technological breakthrough
significantly elevating the success of present and future RNA-targeting drug development projects.
(1) Li, A.; Bouhss, A.; Clément, M.-J.; Bauvais, C.; Taylor, J. P.; Bollot, G.; Pastré, D. Using the Structural Diversity of RNA: Protein Interfaces to Selectively Target RNA with Small Molecules in Cells: Methods and Perspectives. Front. Mol. Biosci. 2023, 10, 1298441. https://doi.org/10.3389/fmolb.2023.1298441
(2) Boca, M.; Kretov, D.; Desforges, B.; Mephon-Gaspard, A.; Curmi, P.; Pastre, D. Probing Protein Interactions in Living Mammalian Cells on a Microtubule Bench. Scientific reports 2015, 5, 17304. https://doi.org/10.1038/srep17304
(3) El Hage, K.; Babault, N.; Maciejak, O.; Desforges, B.; Craveur, P.; Steiner, E.; Rengifo-Gonzalez, J. C.; Henrie, H.; Clement, M.-J.; Joshi, V.; Bouhss, A.; Wang, L.; Bauvais, C.; Pastré, D. Targeting RNA:Protein Interactions with an Integrative Approach Leads to the Identification of Potent YBX1 Inhibitors. eLife 2023, 12, e80387. https://doi.org/10.7554/eLife.80387