Drug R&D productivity declines
The introduction of a new drug to the market is currently associated with a lengthy timeline of approximately 10-15 years, requiring from $1 to $3 billion. Throughout the drug development journey, a therapeutic agent faces a significant 90% probability of failure due to issues related to pharmacokinetics, efficacy, or off-target-associated toxicity [1].
In response to this stringent bottleneck, the pharmaceutical industry continually refines each phase of the drug development process. Thus, recent years have seen remarkable advancements in both computational and biological sciences, with the consequent emergence of new druggable disease triggers waiting for innovative solutions to be targeted. Of particular interest, new roles have been identified for RNAs in cellular functions and gene expression, making RNA targeting, offering promising avenues for restoring processes impaired in various human diseases, including cancer and neurodegenerative disorders. Within this evolving landscape, our Synsight talent team takes advantage of both an innovative biological approach and last artificial intelligence solutions to take up the challenge, with a peculiar focus on RNA targeting. This strategy holds the promise of offering successful perspectives for the discovery of innovative, next-generation medicines in a fast and cost-efficient fashion.
AN INNOVATIVE CELL BASED ASSAY FOR NEW SOLUTIONS
Synsight develops an exclusive, patented technology to detect in cellulo interactions: the Microtubule Bench assay: MTbench®, originated from 15 years of R&D from INSERM (French national institute of health) [2],[3].
The MTbench® assay uses the microtubule network as an intracellular bench to detect protein interactions or RNA-protein interaction in living cells and use the cellular machinery to deploy the assay setup.
Thus, the MTbench® technology enables direct description of RNA-protein or protein-protein interactions in a live cellular context. Moreover, modulation of the interaction by pharmaceutical agents is measurable, enabling high-throughput screening of these interaction modulators (inhibitors or promoters). In the last few years, we developed, optimized, and standardized the MTbench® technology using lab automation and robotics, and enlarged the spectrum of pharmaceutical applications.
MTbench® is easy to set up and allows for direct visualization within single cells. Its automated and robotized system can screen thousands of compounds, providing statistically relevant data through the analysis of thousands of cells, and provides high sensitivity by detecting activity across nanomolar to micromolar concentrations. The MTbench® screening assay was subsequently coupled to series of complementary assays enabling to finely characterize the mode of action of active compounds and exclude undesired activities.
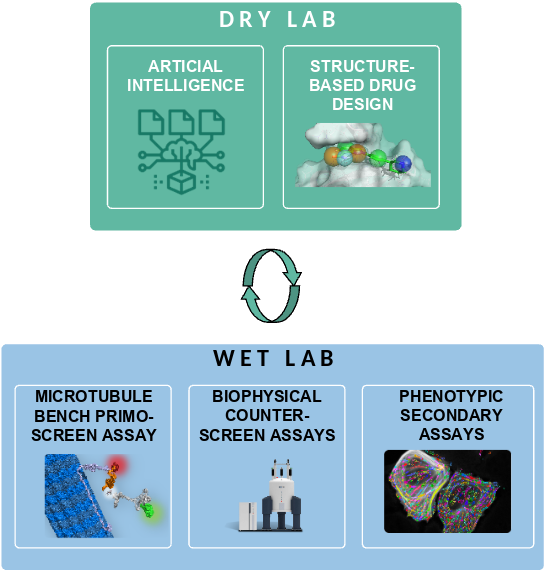
OPTIMIZED ITERATIVE CYCLE
At Synsight, we design small molecules by tackling deep learning, chemoinformatics and data science to develop and implement intelligent algorithms. Our algorithms serve the purpose of swiftly exploring chemical landscapes and, with growing efficiency, identifying bioactive agents suitable for inclusion in RNA-targeting screens. Current target- and ligand-based in silico techniques markedly heightens the likelihood of pinpointing a promising compound within a limited set of tested agents during in cellulo screening [4].
But combining the mentioned techniques with artificial intelligence offers us a great advantage: the capacity –by implementing our iterative selection workflows for ongoing enhancement of our chemoinformatic platform at predicting small molecules activities, as it is continually fed with an increasing volume of robust experimental results from the bench [5]. Hence, our vision is to steadily elevate screening success rates, outperforming conventional drug discovery strategies. This pivotal achievement, applied to our internal initiatives, collaborations, or service projects, founds our rational for accelerated and safer RNA targeting drug discovery.
The resulting screening cascade produces key activity data, from the atomic up to the phenotypic scale. These data are further treated by our computational chemists, who produce in turn an upgraded selection of new compounds injected into our iterative selection workflow.
Conclusion
Our innovative approach combines lab automation, innovative cell-based screening technologies, in silico techniques, and AI-driven insights to improve and accelerate the selection of RNA-targeting drug-like compounds.
We use generative models to design new molecules and virtually screen them before they get tested at the bench in a combination of cell-based and biophysics assays. A key aspect of this approach is that it enables to act on a specific interaction of a specific structural pocket present in any protein of interest, allowing us to finely target key interactions filling stringent druggability criteria. The synergy between our highly skilled scientists from different fields of expertise aims at elevating screening success rates, outperforming conventional drug discovery strategies, and reducing the pharmaceutical attrition rate. Our mission is to bridge the gap between innovative in silico drug design and robust in cellulo screening, ultimately advancing the field of RNA-targeting drug discovery.
By combining cutting-edge technologies and expertise,
we strive to make a significant impact on preclinical cost-efficiency and predictive pharmaceutical investigations,
leading to the development of more effective and safe therapeutic agents for various diseases.
[1] Sun, D.; Gao, W.; Hu, H.; Zhou, S. Why 90% of Clinical Drug Development Fails and How to Improve It? Acta Pharm Sin B 2022, 12 (7), 3049–3062. https://doi.org/10.1016/j.apsb.2022.02.002.
[2] Boca, M.; Kretov, D. A.; Desforges, B.; Mephon-Gaspard, A.; Curmi, P. A.; Pastré, D. Probing Protein Interactions in Living Mammalian Cells on a Microtubule Bench. Sci Rep 2015, 5, 17304. https://doi.org/10.1038/srep17304.
[3] Maucuer, A.; Desforges, B.; Joshi, V.; Boca, M.; Kretov, D. A.; Hamon, L.; Bouhss, A.; Curmi, P. A.; Pastré, D. Microtubules as Platforms for Probing Liquid–Liquid Phase Separation in Cells – Application to RNA-Binding Proteins. J Cell Sci 2018, 131 (11). https://doi.org/10.1242/jcs.214692.
[4] Chang, Y.; Hawkins, B. A.; Du, J. J.; Groundwater, P. W.; Hibbs, D. E.; Lai, F. A Guide to In Silico Drug Design. Pharmaceutics 2023, 15 (1), 49. https://doi.org/10.3390/pharmaceutics15010049.
[5] Han, R.; Yoon, H.; Kim, G.; Lee, H.; Lee, Y. Revolutionizing Medicinal Chemistry: The Application of Artificial Intelligence (AI) in Early Drug Discovery. Pharmaceuticals 2023, 16 (9), 1259. https://doi.org/10.3390/ph16091259.